The platelet-em dataset contains two 3D scanning electron microscope (EM) images of human platelets, as well as instance and semantic segmentations of those two image volumes.
This data has been reviewed by NIBIB, contains no PII or PHI, and is cleared for public release. All files use a multipage uint16 TIF format. A 3D image with size [Z, X, Y] is saved as Z pages of size [X, Y]. Image voxels are approximately 40x10x10 nm3.
Download
Label files can be downloaded as RGBA TIF image volumes or as string-based JSON dictionaries.
TIF
Download images and TIF labels (95.3 MB)
JSON
Download images and JSON labels (98.2 MB)
Image files
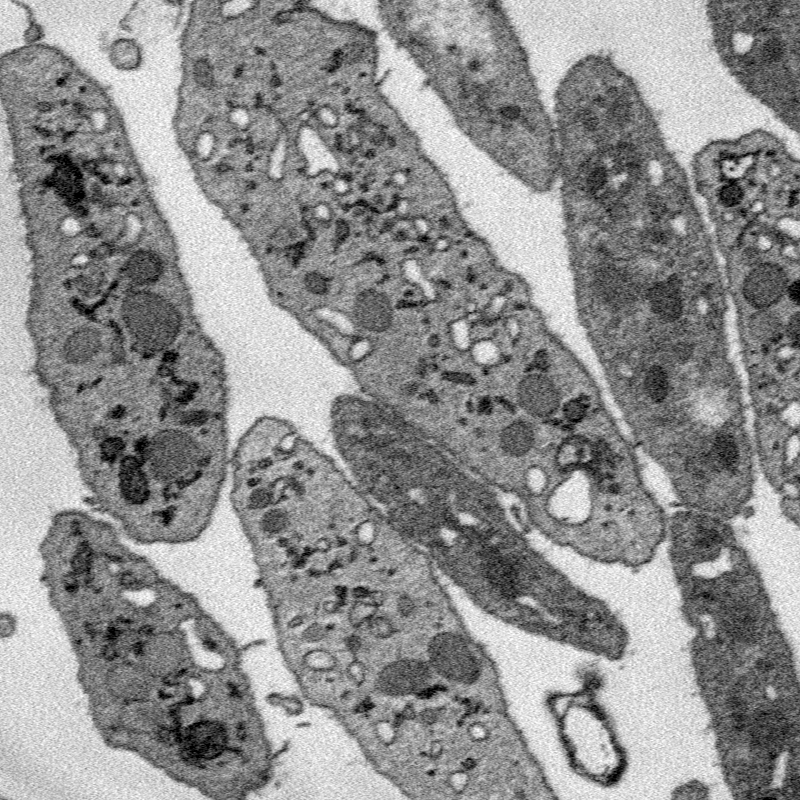
The platelet image volumes were acquired by a Gatan 3View serial block-face scanning electron microscope (SBF-SEM).
Image files are:
-
50-images.tif, a 50x800x800 SBF-SEM image saved as a grayscale TIF.
-
24-images.tif, a 24x800x800 SBF-SEM image saved as a grayscale TIF.
Label files
Label data can be stored in image-based or text-based formats. Image files are better for visualization and image manipulation with GUIs, while text files are better for use with scripting. For its labels, platelet-em dataset uses TIF-formatted image files and JSON-formatted text files.
TIF format
Label TIF files assign a color to each voxel in a corresponding image file. The colors correspond to labels, either object classes for semantic labels or unique object ids for instance labels.
Semantic labels
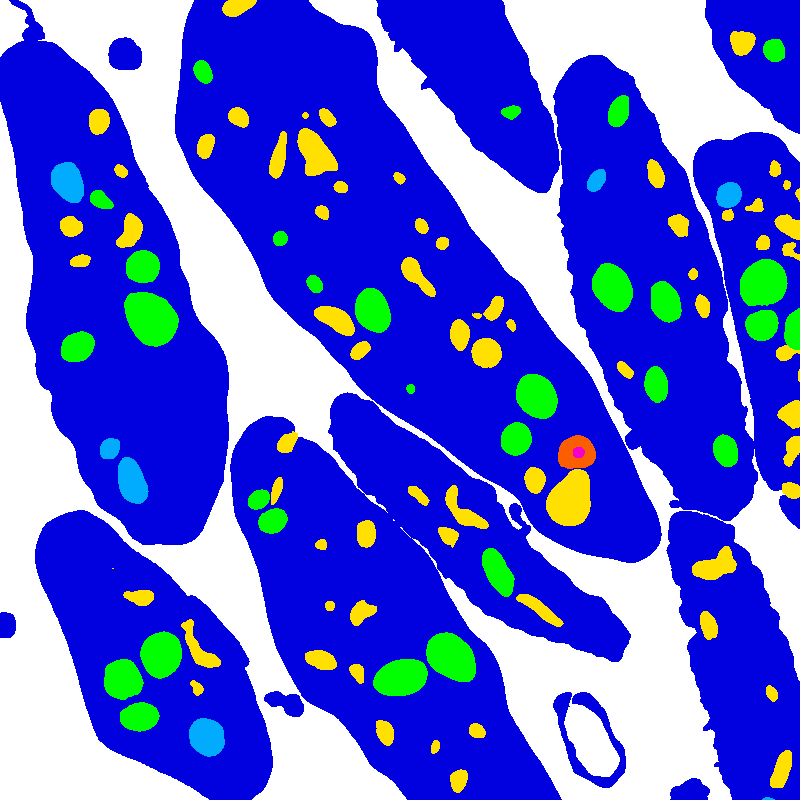
Semantic label files classify each image voxel into one of seven classes, indexed from 0-6:
| Index | Color | Class name |
|---|---|---|
| 0 | None | Background |
| 1 | Dark Blue | Cell |
| 2 | Cyan | Mitochondria |
| 3 | Green | Alpha granule |
| 4 | Yellow | Canalicular vessel |
| 5 | Red | Dense granule body |
| 6 | Purple | Dense granule core |
Semantic label files are:
-
50-semantic.tif: A 50x800x800 semantic segmentation of 50-images.tif, saved as an RGB TIF.
-
24-semantic.tif: A 24x800x800 semantic segmentation of 24-images.tif, saved as an RGB TIF.
Instance labels

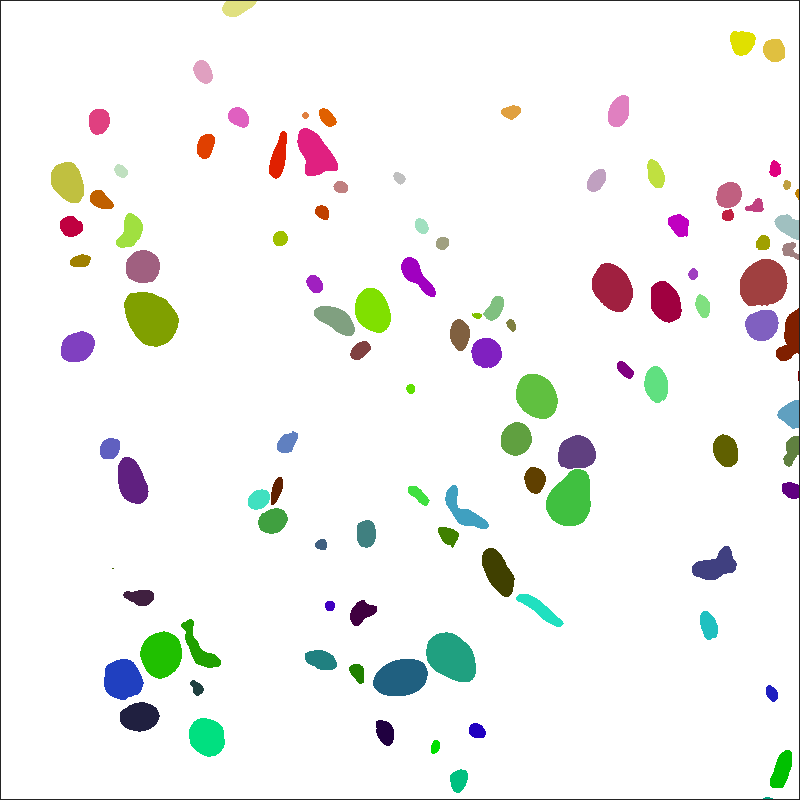
Instance labels are separated into two label files per image: cell and organelle. Cell instance labels assign a unique id to each cell in an image volume. Organelle instance labels assign a unique id to each organelle in an image volume.
IDs are assigned to distinct objects within the entire 3D image volume. Tags for cross-sections of the same 3D object are the same between different 2D image windows.
Instance label files are:
-
50-instance-cell.tif: A 50x800x800 instance segmentation of cells in 50-images.tif, saved as an RGB TIF.
-
50-instance-organelle.tif: A 50x800x800 instance segmentation of organelles in 50-images.tif, saved as an RGB TIF.
-
24-instance-cell.tif: A 24x800x800 instance segmentation of cells in 24-images.tif, saved as an RGB TIF.
-
24-instance-organelle.tif: A 24x800x800 instance segmentation of organelles in 24-images.tif, saved as an RGB TIF.
JSON format
For both semantic and instance labels, label data can be represented with a dictionary structure. Dictionary keys are the unique non-zero integer labels in the label data. For each key, the corresponding value is the binary mask of the region associated to the key. This mask is stored as a run-length encoded string. An additional '.info' key stores image shape and datatype information.
File names are the same as for the TIF format, with the .json file format instead of .tif. Functions for converting between TIF and JSON formats, as well as between python strings and numpy ndarrays, can be found in the bio3d_vision package.
Example
In python - a synthetic blob image with two nonzero values:
blob1 = skimage.data.binary_blobs(length=10, seed=1, volume_fraction=0.2)
blob2 = skimage.data.binary_blobs(length=10, seed=2, volume_fraction=0.1)
image = np.zeros((10, 10), dtype=int)
image[blob1] = 1
image[blob2] = 2
has the following JSON-compatible dictionary representation:
image_as_dict = {
'.info': {'dtype': 'np.int', 'shape': (10, 10)},
'1': '7 2 12 1 21 1 40 1 50 4 60 3 68 1 71 1 81 1',
'2': '1 3 11 1 33 2 42 3 87 2'}