The bio3d-vision dataset collection's goal is to aggregate high-quality, large-scale 3D biological microscopy datasets whose analyses present challenging computer vision problems that are important to advancing the state of microscopy.
The bio3d-vision project is organized by LCIMB, a part of NIH/NIBIB.
Getting started
In addition to manual dataset downloads below, this project provides a bio3d_vision python package to simplify downloading and manipulating 3D microscopy images and labels.
Requirements
The bio3d_vision package uses python 3.6+ features, and requires matplotlib, numpy, scipy, scikit-image, and tifffile.
Clone the repo
Clone the bio3d_vision package from https://github.com/bio3d-vision/bio3d_vision. Add it to your system path or a project folder to use it. TODO: pip integration.
Example: platelet-em
Click here for an example of using bio3d_vision to download the platelet-em dataset and load it into numpy for manipulation and windowing.
Datasets
platelet-em
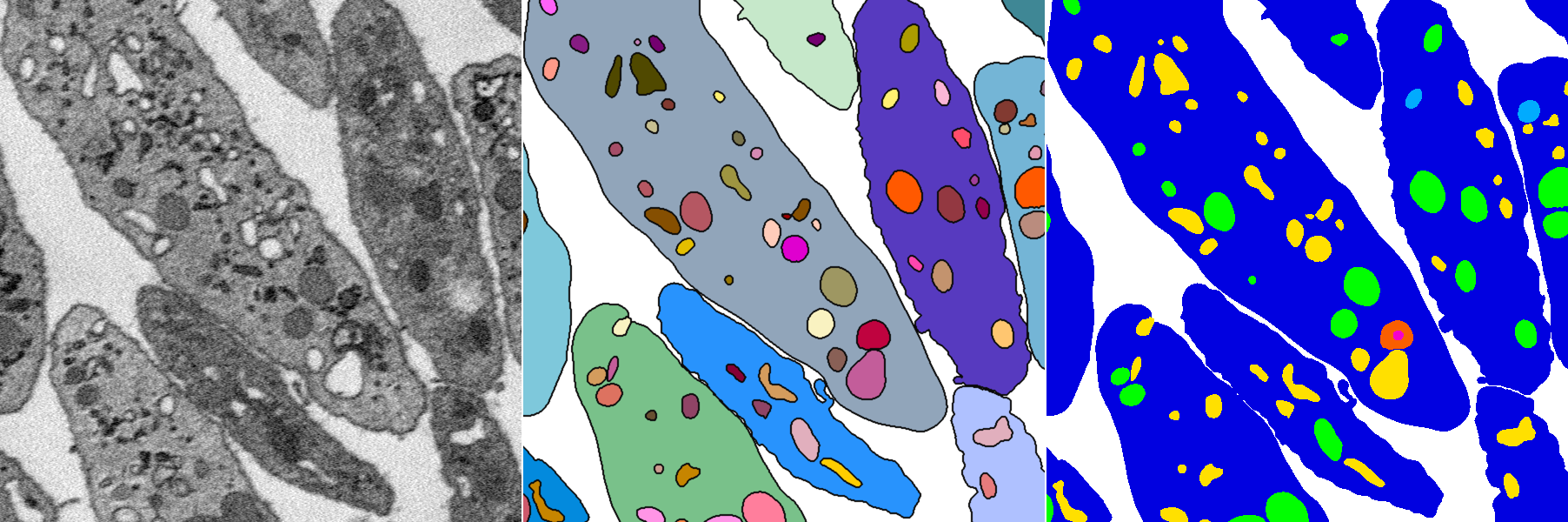

Created by LCIMB in collaboration with Prof. Brian Storrie's lab at UAMS.
Download
Label files can be downloaded as RGBA TIF image volumes or as string-based JSON dictionaries. The TIF format is better for visualization and image manipulation GUIs, the JSON format is better for use with scripting.
TIF
Download images and TIF labels (95.3 MB)
JSON
Download images and JSON labels (98.2 MB)
Description
Click here for a detailed description of the platelet-em dataset.